
Overview of Development in Machine Learning and Meta-genomic Techniques to Discover the Prognosis Tool for Parkinson’s Diseases.
Neurodevelopmental diseases have garnered strong association with gut microbial environment in recent development of our understanding of gut-brain axis and intestinal permeability(Wallen et. al,). The drastic changes in microbial landscape and the concentration of microbial byproducts alter the onset of Parkinson's disease(PD) in susceptible patients. Machine Learning(ML) based meta analysis reveals gut microbiome alternation shows a pattern in PD patients. Authors suggest their novel training model could account for varying degrees of unaccounted factors such as age, sex, diet, treatment, culture from the supplied data. The lack of prior meta analysis to fall back on, the novel ML techniques used in these studies validates its results in its alignment with epidemiological evidence. Factors analysed in these studies are the microbiome diversity and presence of pathogenic biocursors like Short chain fatty Acids(SFCAs) and Butyrate compounds.
Abstract
Neurodevelopmental diseases have garnered strong association with gut microbial environment in recent development of our understanding of gut-brain axis and intestinal permeability(Wallen et. al,). The drastic changes in microbial landscape and the concentration of microbial byproducts alter the onset of Parkinson's disease(PD) in susceptible patients. Machine Learning(ML) based meta analysis reveals gut microbiome alternation shows a pattern in PD patients. Authors suggest their novel training model could account for varying degrees of unaccounted factors such as age, sex, diet, treatment, culture from the supplied data. The lack of prior meta analysis to fall back on, the novel ML techniques used in these studies validates its results in its alignment with epidemiological evidence. Factors analyzed in these studies are the microbiome diversity and presence of pathogenic biocursors like Short chain fatty Acids(SFCAs) and Butyrate compounds.
Highlights
- Gut microbiome differs significantly between PD patients and control, measures of microbial alpha-diversity and abundances of rare taxa, based on species profiles, were higher in PD samples.
- Genera Lactobacillus, akkermanisa, and Bifidobacterium were abundant in PD samples.
- Short chain fatty acids producing bacteria from the Lachnospiraceae family and Faecalibacterium genus were found depleting in PD patients.
- Microbiome-based machine learning(ML) accurately classifies PD patients with average AUC 71.9% in study specific models. However, the model did not generalise well across other studies(AUC 61%)
- Training Models on multiple datasets improves their generalizability( LOSO AUC 68%)
- Meta analysis of shotgun metagenomics portrays PD-associated microbial pathways such as increased microbial pathways for solvent and pesticide biotransformation.
Introduction
Parkinson’s Disease(PD) is the second most common neurodegenerative disorder. Due to the progressive aging of the population, PD is expected to be doubled by 2030. PD affects the dopaminergic neurons in the brain. The subsequent loss of dopamine leads to motor impairments, such as tremor, rigidity, balance difficulties, and loss of spontaneous movements. Pathologically, intercellular deposition of aggregated and misfolded apla-synuclein(a-syn) leads to neuronal cell death and neuro inflammation. Pathogenicity affects the both peripheral and central nervous system of the body causing non-motor symptoms often gastrointestinal irregularities. These irregularities often precede the impending motor symptoms for years. Through Gut brain axis, a bidirectional neurological and chemical tract between the brain and intestine, onset of these gastrointestinal irregularities can onset or trigger the motor impairment often seen in PD patients. Thus studying how the changes in gut influences the brain and what changes in particular bring about the motor impairment in PD patients is of vital importance. This paper explores the recent studies in gut microbiota, PD patients, meta-genomics and machine learning application to understand and find the correlation and pattern of changes associated in PD to aid in the development of standardized panel to diagnose PD symptoms before the onset of motor-impairments which can prevent the severity of PD due to early intervention in genetically predisposed and likely PD candidates.
- Dysbiosis of the Gut, difference between PD and control samples.
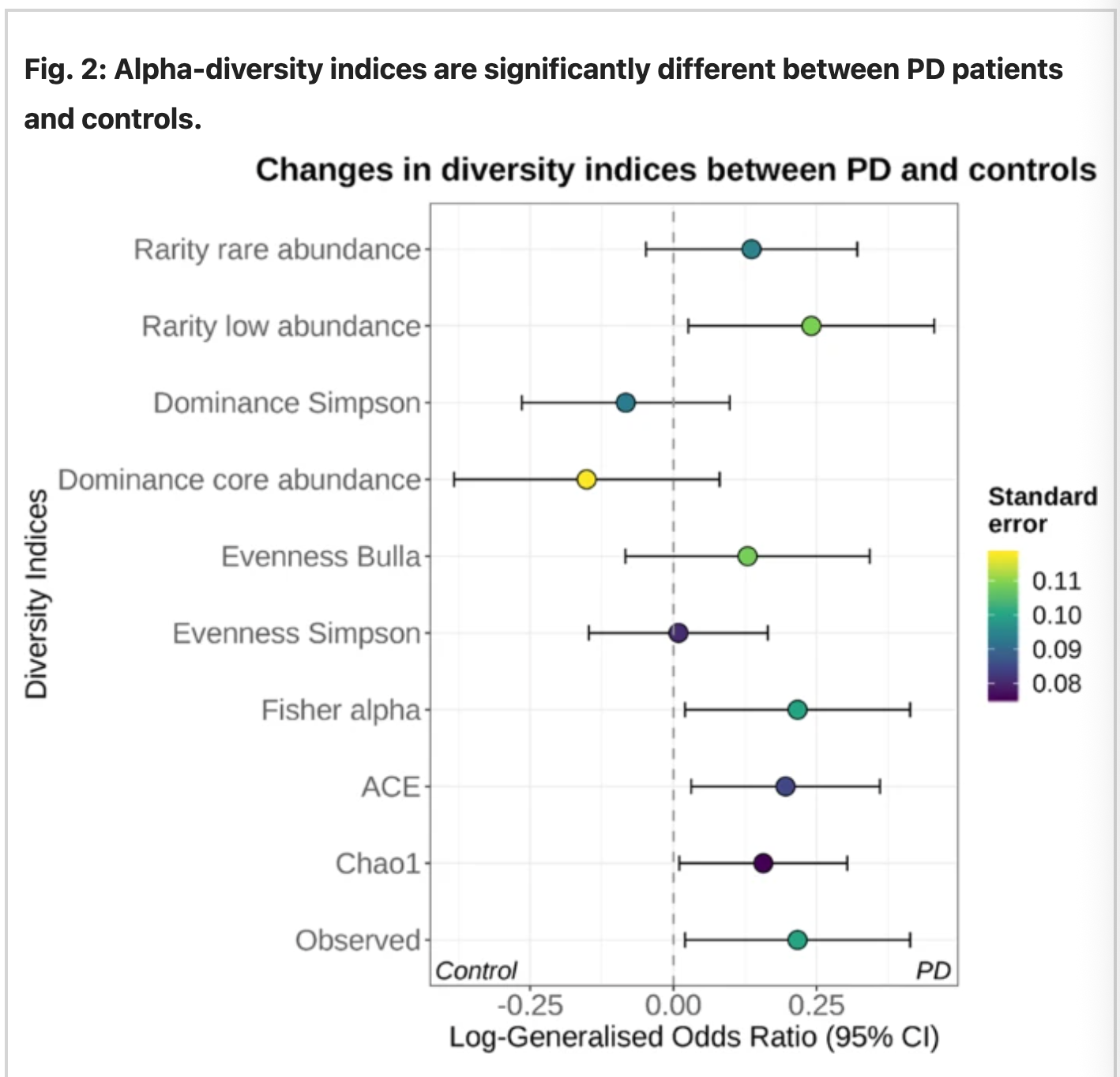
Source: Nature
Fig 1: Increased diversity of rare genus in PD sample.
The species level for each dataset was combined using random-effect-meta-analysis. The abundance scale was determined using the observed number of species and the indices Chao1, ACE, and Fisher’s alpha. The figure above indicates the presence of rare genre bacteria to be present abundantly in PD samples than control.
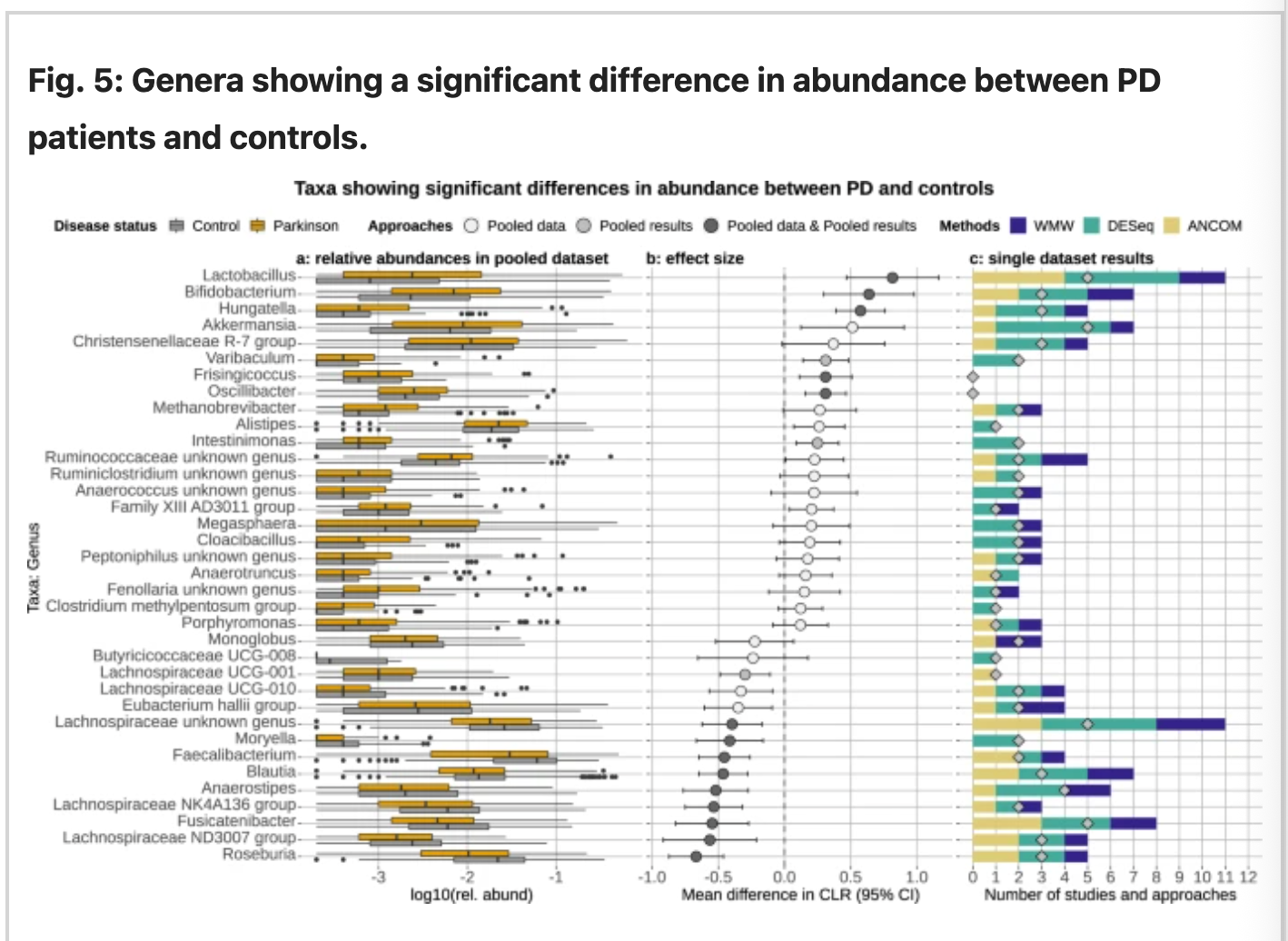
Source: Nature
Fig 2: the relative abundance of the genera retrieved from the rarefield pooled data reported in a, estimated sizes and b, effect sizes and c, the number time each genus detected.
The genus specified table from figure 2 suggests the relative abundance of Roseburia, Blautia, Fusicatenibactor, Faecalibacterium, Moryella, Anaeorostipes, from family Lachnospiraceae in control. In contrast, PD samples were abundant in the genera Lactobacillus, Bifidobacterium, Hungatella, and Akkermansia and few groups from the CHristensenellaceae family. The testing between predicting model and taxonomical data suggests that majority of predicted pathways for PD were related to ubiquinone(Coenzyme Q; CoQ) and Menaquinone biosynthesis, glutamate degradation, methanogenesis, and lactic-type fermentation. While for the control sample, taxonomical data relay the increased biological pathways involved in biosynthesis of Vitamin B12 and glutamine/mate, degradation of glucuronate and galactoglucuronate, and methane production. (Romano et al., 2021)
Bedarf results from 2017 found the Verrucomicrobiaceae, Firmicutes and Prevotellaceaceae and Erysipelotricheceae were significantly altered in PD patients which could be distinguished from the control with ROC-AUC of 0.84. (Bedart et. al, 2017) These findings were consistent with PD involved in the intestinal barrier function and immune response. Furthermore, Keshavarzian's paper suggests where PD patients' samples were correlated with alpha-synuclein aggregation in their colon with evidence suggesting intestinal inflammation. The paper highlights the abundance and decline of abovementioned taxa contributing to the dysbiosis which could trigger the misfolding of a-syn. (Keshavarzian et. al. 2015)
- ML analysis shows the alteration of Microbial communities in PD control
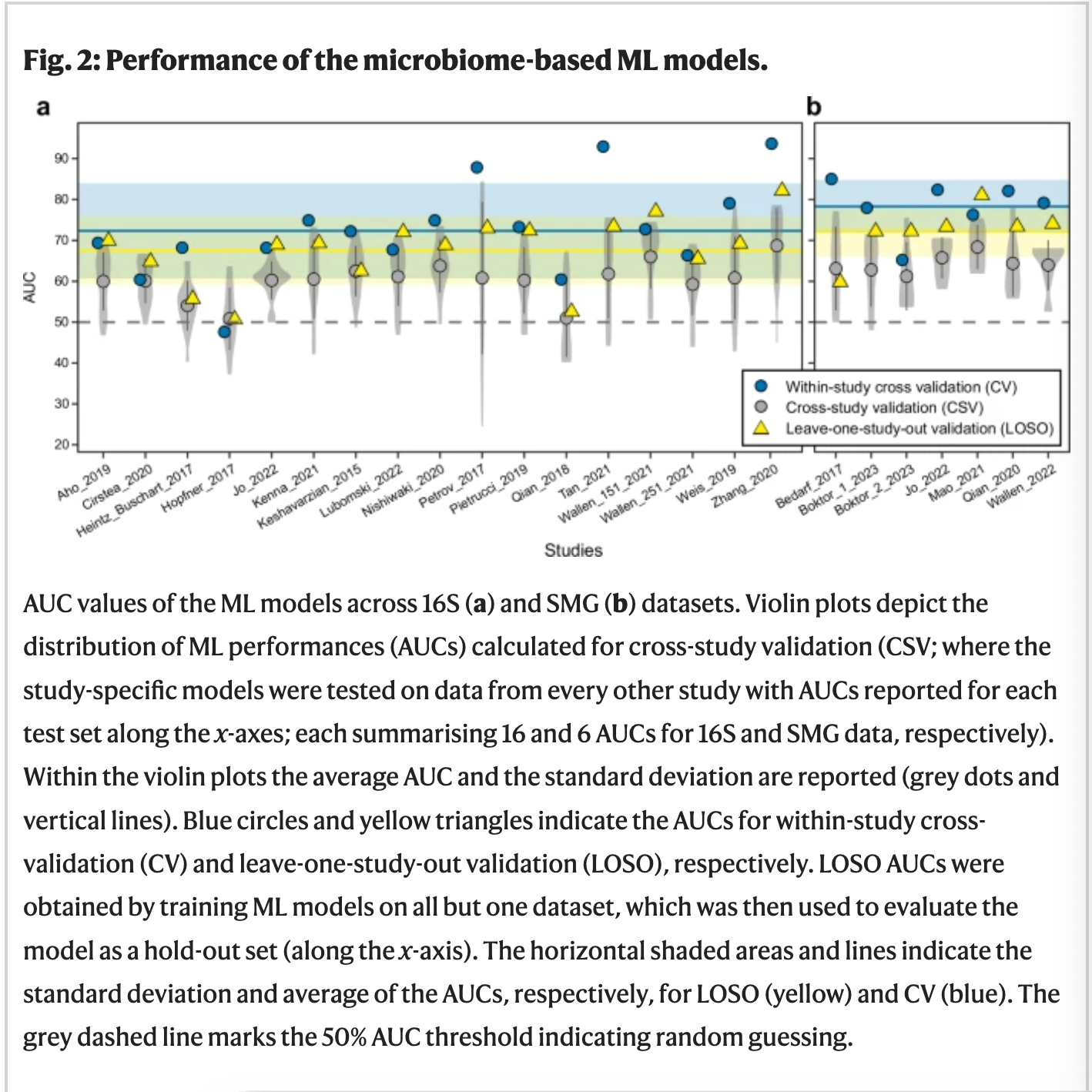
Figure 3: ML model performance
4489 data of fecal samples from PD patients were combined from 22 case-controlled studies from 4 continents. The microbiome profile from sample and control was attained using 16S amplicon and shotgun metagenomics sequencing(SMG). ML models were designed by filtering strategies, normalisation approaches, and ML algorithms in R package SIAMCAT. Area under the receiver operating characteristics curve(AUC) was used to evaluate the accuracy of the model. The models were evaluated for both 16S and SMG datasets.
The models performed well within study cross validation(CV) while some dataset performed better than others. The models tested for one study were cross examined(CSV)for another to examine the prediction accuracies when tested on independent datasets. Compared to the higher AUC for case specific model, cross-study portability of ML model declines. However, when the study was performed on a single subset of species, accuracies were again higher. The cross-disease specificity of the model, the tendency of the model to wrongly predict patients affected by other neurodegenerative diseases, were assessed by testing PD models on data obtained from studies investigating other diseases. For Alzheimer's and Multiple Sclerosis cross studies, the accuracies of the ML model greatly varied from 0-100% averaging at 35.1%. In the same study, when ML models were replaced by (leave one study out) LOSO models, the prediction drastically improved.(Romano et al., 2025)
Discussion
In comprehensive analysis of datasets used in these models and studies were derived from 16S and SMG. The in-vivo data used in the models are limited with 4489 data being the highest pool of dataset ever used. The ML model accuracy was severely depleted due to limited cross-compatibility of the datasets. The novel models created for the metagenomic studies using LOSO validation, CV, CSV are crucial developments to improve the accuracy and model practicality of the models. This helps the novel models to cross-validates its accuracies through datasets from various independent studies. LOSO in particular, generalizes the datasets filtering out the study based biases before being fetched into the model.
Neural networks and large language models can radically improve the outcome of the prediction Models in the near future. However, the field needs the crude supply of datasets from PD patients and control to test these large models. Alphafold used X-Ray crystallography, NMR spectroscopy, and Cryo-EM data to validate its prediction accuracy. Alphafold also uses the standardized data available from thousands of independently annotated models from Protein Data Bank to validate its prediction accuracy. In contrast, the metagenomics of gut-microbiome lacks the experimental prognosis techniques for PD and other neurodevelopmental disorders similar to PD. Similarly it also lacks the central database of in-vivo genomic datasets including, microbial profile of the gut, inflammatory or permeability score of gut lining, involvement of medication and other crucial factors to validate its models.
Gut microbiome is home to millions of bacterial and archeal species and their composition is greatly influenced by sex, age, diet, and other regional variation.Inorder filter unassociated factor out LOSO validation model needs to have to advance understanding of formation and molecular precursor related to the formation of microbiome diversity. Depleting amounts of SFCAs and butyrate compounds are now established as key factors associated with membrane permeability which helps the pathogenic molecule to translocate to the brain and vice versa. Lateral development of the gut-brain axis is also crucial in these regards and it directly helps in the development of new models regulating and monitoring biomarkers. Neurodegenerative disorders are difficult to reverse once the neural damage in the brain is triggered, these damages change the gut-biome which further accelerates the degenerative process of brain functions. Treatment of neurological disorder can be drastically improved with early intervention delaying the onset of irreversible neurological symptoms and tools like ML are cheap and effective ways to monitor the patients.
**All the published code and algorithm used in the study are available for public use on nature.com
Useful Links:
References
Bedarf, J. R. et al. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-na‹ve Parkinson's disease patients. Genome Med. 9, 1–13 (2017).
Cersosimo, M. G., & Benarroch, E. E. (2012). Pathological correlates of gastrointestinal dysfunction in Parkinson’s disease. Neurobiology of Disease, 46(3), 559–564. https://doi.org/10.1016/j.nbd.2011.10.014
Clairembault, T., Leclair-Visonneau, L., Coron, E., Bourreille, A., Le Dily, S., Vavasseur, F., Heymann, M.-F., Neunlist, M., & Derkinderen, P. (2015). Structural alterations of the intestinal epithelial barrier in Parkinson’s disease. Acta Neuropathologica Communications, 3(1). https://doi.org/10.1186/s40478-015-0196-0
Dauer, W., & Przedborski, S. (2003). Parkinson’s Disease: Mechanisms and Models. Neuron, 39(6), 889–909. https://doi.org/10.1016/s0896-6273(03)00568-3
Elbaz, A., Carcaillon, L., Kab, S., & Moisan, F. (2016). Epidemiology of Parkinson’s disease. Revue Neurologique, 172(1), 14–26. https://doi.org/10.1016/j.neurol.2015.09.012
Keshavarzian, A. et al. Colonic bacterial composition in Parkinson’s disease: colonic microbiota in Parkinson’s disease. Mov. Disord. 30, 1351–1360 (2015).
Nishiwaki, H., Ito, M., Ishida, T., Hamaguchi, T., Maeda, T., Kashihara, K., Tsuboi, Y., Ueyama, J., Shimamura, T., Mori, H., Kurokawa, K., Katsuno, M., Hirayama, M., & Ohno, K. (2020). Meta‐Analysis of Gut Dysbiosis in Parkinson’s Disease. Movement Disorders, 35(9), 1626–1635. https://doi.org/10.1002/mds.28119
**Romano, S., Jakob Wirbel, Ansorge, R., Schudoma, C., Quinten Raymond Ducarmon, Arjan Narbad, & Zeller, G. (2025). Machine learning-based meta-analysis reveals gut microbiome alterations associated with Parkinson’s disease. Nature Communications, 16(1). https://doi.org/10.1038/s41467-025-56829-3
**Romano, S., Savva, G. M., Bedarf, J. R., Charles, I. G., Hildebrand, F., & Narbad, A. (2021). Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. Npj Parkinson’s Disease, 7(1), 1–13. https://doi.org/10.1038/s41531-021-00156-z
Schwiertz, A., Spiegel, J., Dillmann, U., Grundmann, D., Bürmann, J., Faßbender, K., Schäfer, K.-H., & Unger, M. M. (2018). Fecal markers of intestinal inflammation and intestinal permeability are elevated in Parkinson’s disease. Parkinsonism & Related Disorders, 50, 104–107. https://doi.org/10.1016/j.parkreldis.2018.02.022
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermudez-Humaran, L. G., Gratadoux, J.-J. ., Blugeon, S., Bridonneau, C., Furet, J.-P. ., Corthier, G., Grangette, C., Vasquez, N., Pochart, P., Trugnan, G., Thomas, G., Blottiere, H. M., Dore, J., Marteau, P., Seksik, P., & Langella, P. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences, 105(43), 16731–16736. https://doi.org/10.1073/pnas.0804812105
Wallen, Z. D., Demirkan, A., Twa, G., Cohen, G., Dean, M. N., Standaert, D. G., Sampson, T. R., & Payami, H. (2022). Metagenomics of Parkinson’s disease implicates the gut microbiome in multiple disease mechanisms. Nature Communications, 13(1). https://doi.org/10.1038/s41467-022-34667-x
Wirbel, J., Zych, K., Essex, M., Karcher, N., Kartal, E., Salazar, G., Bork, P., Sunagawa, S., & Zeller, G. (2021). Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox. Genome Biology, 22(1). https://doi.org/10.1186/s13059-021-02306-1



